From Discussions VOL. 5 NO. 2The Role of Innate Receptor TLR2 in Neutrophil Recruitment in Oropharyngeal Candidiasis (OPC)
IN THIS ARTICLE
KEYWORDS
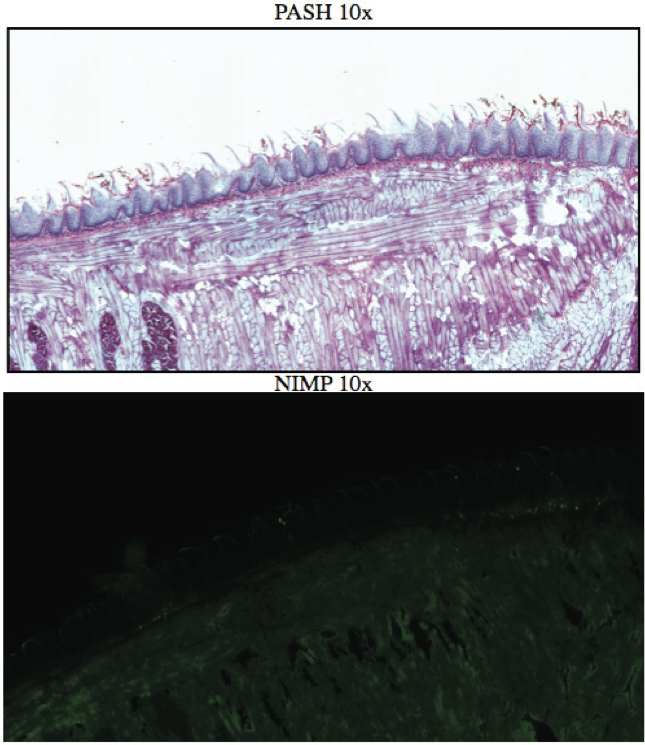
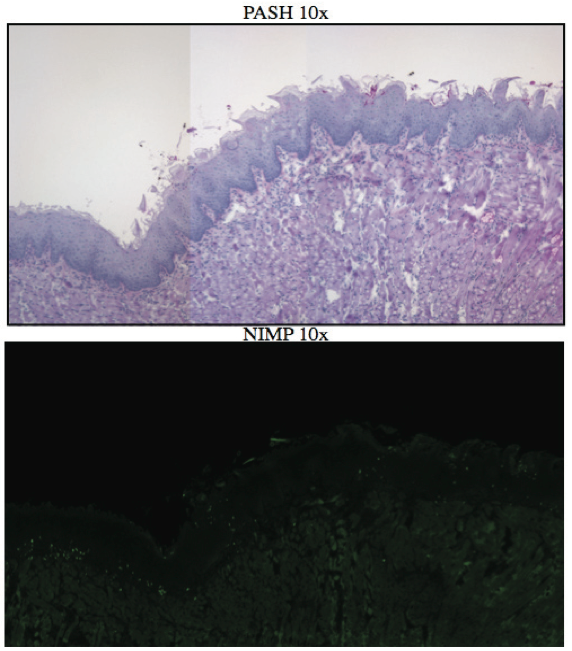
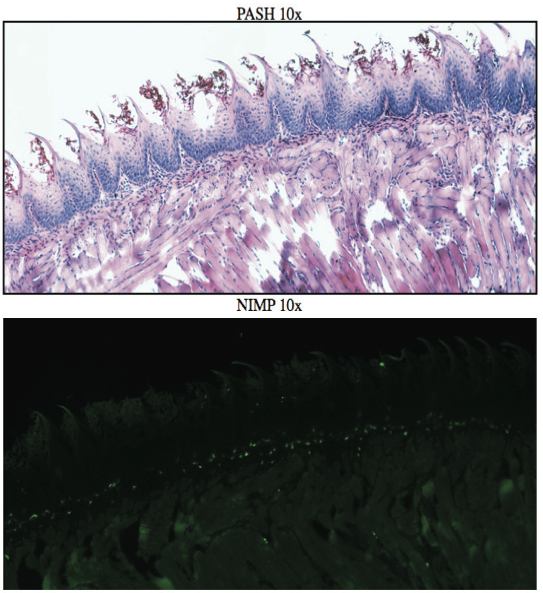
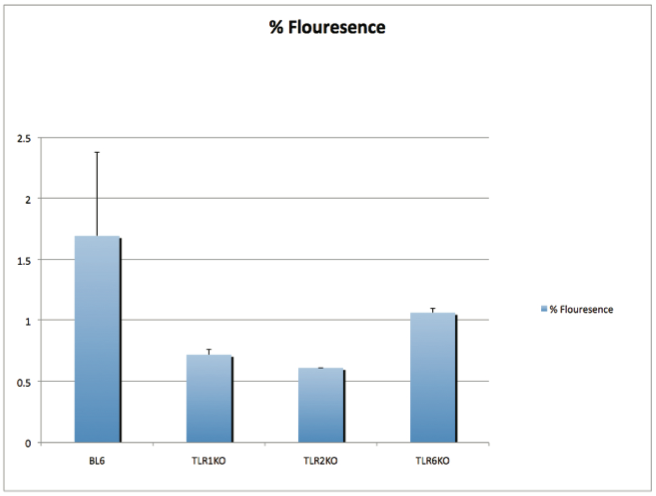
AbstractCandida albicans (CA) is a dimorphic fungus that commonly colonizes the oral cavity. Overgrowth can result in an infection of the oral cavity, known as oral candidiasis (OPC). Neutrophils play a major role in nonspecific immune defense against fungal infection. Neutrophils then engage in phagocytosis of the C. albicans by engagement of membrane innate receptors, called pattern recognition receptors (PRRs) that interact with certain identifying molecules on the cell wall of the infecting fungus, called pathogen-associated molecular patterns (PAMPs). Toll Like Receptors (TLRs) are a type of PPR expressed on neutrophils and are believed to play a major role in its activation and migration. This study evaluates the role of TLR1, 2, and 6 in neutrophil recruitment in tongues utilizing a murine model of OPC. Tongue sections from C57BL/6, TLR1, TLR2, and TLR6 KO mice were stained with a neutrophil specific NIMP/alex488 antibody. Neutrophil recruitment was quantitatively assessed by comparing the amount of fluorescence in the tongue epithelium. Our results found that the lowest percent fluorescence was experienced in the tongue sections of the TLR2KO mice meaning that neutrophil recruitment was hindered. We also observed hindered neutrophil recruitment in the TLR1, and TLR6 KO mice, however it was not as severe as in the TLR2 KO mice. These results imply that all three TLR types 1, 2, and 6 play a role in neutrophil recruitment in the OPC model. IntroductionThe human oral cavity serves as a reservoir for hundreds of commensal microbes that coexist within a delicate balance, between microbes and the host immune system. These microbes live together in complex networks termed biofilms, in which the microbes interact cooperatively to promote survival and growth. Many parameters affect this biofilm such as temperature, pH, oxygen, fluid flow, as well as the host's age. In particular, the tongue offers microbes an extremely fertile environment for biofilm colonization, due to its numerous protected sites.1 Any disturbance in the oral cavity's normal flora can allow for the overgrowth of normal commensal organisms, such as Candida albicans, resulting in a pathogenic infection. The propagation of this fungus can result in a very painful and in some cases fatal condition known as oropharyngeal candidiasis (OPC).2 Candidiasis is an opportunistic pathogen, meaning that it usually does not cause disease in healthy individuals. Hence, hosts who are immunocomprimized; typically elderly, those who suffer from AIDS, people that undergo chemotherapy, infants, or transplant patients, are at higher risk of infection. CA is the 4th most common life threatening infection among all bloodstream and nosocomial infections in the United States.3 Candida species are dimorphic fungi capable of existing in both a yeast form and a hyphal form. One of the complicating factors of Candida infection is its ability to switch morphogenic states at body temperature. Yeast and hyphae express different surface molecules at different levels making the initiation of effective immune responses to infection more difficult.4 When invading tissue, Candida is most commonly found in the hyphal form, which is capable of destroying surrounding tissue via mechanical processes. The first line of defense against infection involves the physical barrier provided by epithelial cells. Once Candida has broken through the barrier of the epithelia, control of infection and prevention of dissemination involves the immediate activity of the innate immune system. The main cells of the innate immune system that recognize invading pathogens are monocytes and neutrophils. Neutrophils are polymorphonuclear, phagocytic cells that are part of the innate immune system's first responders to a pathogen infection. Upon infection, a variety of chemical mediators, called cytokines and chemokines, are released resulting in the increased surface expression of adhesion molecules involved in neutrophil infiltration and trigger dilation in the blood vessels around the infected area. This in turn causes added permeability of blood vessels and endothelial cells. Neutrophils then migrate from capillaries and pass through the endothelial layer to the site of infection. This migration from the blood to the site of infection utilizes a process known as chemotaxis. Chemotaxis involves the movement of neutrophils from areas of low chemokine concentration, such as blood, to areas of high chemokine concentration, the site of Candida infection. There is a multitude of protein and carbohydrate molecules that play a critical role in the migration of neutrophils to sites of infection. Until recently, little was known about the process by which neutrophils recognize C. albicans as a pathogen. The oversimplified belief that innate immunity was nonspecific and employs an 'ingest and destroy' method could not explain the specific and diverse responses of innate immunity. Current research shows that pattern recognition receptors (PRRs) assume the role of recognizing, for the innate immune system, invading pathogens including C. albicans. PRRs recognize conserved molecular motifs within pathogens called pathogen-associated molecular patterns (PAMPs). In mammalian molecules, four types of PRRs have been identified as Toll-like receptors (TLRs), C-type lectin-like receptors, Nucleotide-binding oligomerization domain (NOD) like receptors, and retinoic-acidinducible gene receptors. TLRs are transmembrane proteins containing an extracellular domain to identify PAMPs and an intracellular domain that activates signaling cascades upon PAMP interaction. Currently, 10 TLRs have been identified in humans that are capable of engaging specific PAMPs. An example is TLR2, which recognizes phospholipomannan, a phylogenetically unique fungal component. C-type lectin-like receptors mainly constitute membrane bound receptors that recognize polysaccharide structures, such as β-glucans, that are found in the cell wall of C. albicans5. Nucleotide-binding oligomerization domain (NOD)-like receptors and retinoic-acid-inducible gene receptors are both cytoplasmic receptors, involved in recognizing viral nucleic acids. No studies have documented any involvement of these receptors in the recognition of fungal pathogen components.6 Research utilizing fluorescent analysis of neutrophils stained with anti-TLR antibodies has indicated that neutrophils have been shown to express all known TLRs except for TLR37,8. These TLRs play a role in the recognition of the components of fungal pathogens, and further activation and recruitment of neutrophils9. The aim of this research is to investigate neutrophil recruitment into the epithelial tissue of the tongue in response to oropharyngeal candidiasis (OPC) and its dependence on TLR2 and its co-receptors TLR1 and TLR6. Due to the ability of TLR2 to recognize fungal PAMPs, we expect to see a much lower neutrophil count in tongue sections of TLR2KO mice. Doing this will give us a better understanding of the specific roles of TLRs 1, 2, and 6 in the recognition of C. albicans for neutrophil recruitment, and further help us understand the mechanisms that propagate the innate immune response. Materials & MethodsAnimalsC57BL/6 mice were used as the control for this experiment. They were obtained from Jackson Laboratories (Bar Harbor, ME). TLR2KO, TLR6KO, TLR1KO mice were graciously provided by Dr. Shizuo Akira (Osaka University, Osaka, Japan). All animals used in this experiment were housed in filter-covered micro-isolator cages. During breeding and storing they were housed in the Wolstein Animal Facility at Case Western Reserve University. Mice were then transferred to the Wearn Animal Facility at University Hospitals in Cleveland for infection and the duration of the experiment. Case Western Reserve University School of Medicine Institutional Animal Care and Use Committee has approved all protocols and experiments. Fungal preparationThe strain of C. albicans (CA) that was utilized in this study was GDH2346, originally isolated from a denture stomatitis patient. The strain was propagated and maintained in Sabourand Dextrose (SD) agar at 4 ̊C. Prior to mouse infection C. albicans was scraped off the SD agar and transferred to a yeast-inducing SD broth (Difco, Sparks, MD) and incubated at 37oC with a shaking for a period of 16 – 24 hrs. After incubation, yeast was collected and washed 3x with PBS. Yeast was spun down and pelleted using a centrifuge between each wash step. The CA was then suspended in PBS and cells were counted on a hemacytometer. Yeast was then diluted to the desired concentration of 5x107 yeast/mL OPC infection ModelFive days before infection with C. albicans, the mice were fed tetracycline water (2.5g/L), a form of antibiotic, to reduce competing oral flora. The mice were then injected with an anesthesia cocktail (25% Ketamin (Vedco, St Joseph, MO), 7% Acepromazine (Boehringer Ingelheim, Germany). After the mice had been properly anesthetized, 4-6 evenly spaced incisions were made on the dorsal layer of the tongue making sure not to pass the epithelium and cause bleeding. All incisions were made using a sterile #10 blade scalpel. An autoclaved cotton ball saturated with PBS was then inserted into the mouse's mouth to prevent dryness. After 3 hours the cotton ball was removed and 100 μL of 5.0 x 107 CA suspension was pipetted onto the tongue with a new cotton ball, and the animal was left for approximately 4-6 hours while sedated. After a three day infection period the mice were then euthanized using a CO2 asphyxiation chamber. Using sterile autoclaved instruments the tongue was removed and treated for slide preparation. Three different BL6 mouse tongues were obtained as well as two from each of the TLR1KO and TLR6KO mice, and finally only one TLR2KO mouse tongue was obtained. ImmunohistochemistryAfter the tongues were removed they were immersed in Embedding Media (Tissue Tek) (EMS Hatfield, PA) and flash frozen using liquid nitrogen. The tongues were then transferred on dry ice to a microtome-cryostat to be sectioned. From each tongue 2 slides containing two 5-micron sections each, and one slide containing two 10-micron sections were prepared. All slides were then stored in a -20°C refrigerator. The 10-micron slide was stained with Periodic acid-Schiff and hematoxylin (PASH). One of the 5-micron slides was stained with an anti-neutrophil antibody (NIMP supernatant) and incubated for 2 hours. After washing with PBS-triton the slide was then stained and incubated for 45 minutes with the secondary antibody conjugated to the green fluorochrome Alexa-488 at a dilution of 1:400 in 1% normal goat serum, which provided the fluorescent tag that allowed neutrophil detection and imaging with the florescent microscope. After washing again with PBS-triton the slide was then mounted in Vectashield containing the nuclear stain DAPI (Vector Laboratories, Burlingame, CA). The other 5-micron slide was used as the negative control for the neutrophil staining. This was done by using the same method for staining the first 5-micron slide without using the primary antibody (NIMP). Microcopy and QuantificationThe images were taken using a Leica DMI 6000B inverted microscope. For each NIMP slide, a 10x resolution image was taken of the whole tongue at an exposure of 4500 milliseconds. This was accomplished by stitching together a large number of consecutive 10x images of the tongue. In addition to the stitched image a 20x image was taken of the area of high fluorescence both in green fluorescence (Alex488) as well as a combined superimposed image of green and blue fluorescence (Alexa488 & DAPI). For the PASH stained slides a 10x stitched image of the whole slide was taken as well as 20x images of the area of the epithelium where fungal hyphal infiltration was observed. To quantify the number of neutrophils in each tongue section, the cell counting technique was opted for digital quantifying method using the imaging program Metamorph. For each image taken by the program a number between 1 and 4095 is assigned for each pixel corresponding to its "fluorescing" value. To quantify the results we used the 10x-stitched images of each tongue and using Metamorph the epithelial region of the tongue was selected. In the selected region the total amount of pixels were counted as well as the total amount of pixels that had number values that was considered fluorescing, which was set between 715 and 4095. The percent fluorescence was calculated by dividing the number of fluorescing pixels by the total amount of pixels in the epithelium.Continued on Next Page » Suggested Reading from Inquiries Journal
Inquiries Journal provides undergraduate and graduate students around the world a platform for the wide dissemination of academic work over a range of core disciplines. Representing the work of students from hundreds of institutions around the globe, Inquiries Journal's large database of academic articles is completely free. Learn more | Blog | Submit Latest in Biology |